Treadmilling motion of dividing bacteria
Scientists reveal the treadmilling motion of dividing bacteria
Published on: 16 February 2017
An international team of scientists using the latest imaging techniques have revealed how bacterial division proteins build a partition wall across the cell, one molecule at a time.
Publishing in Science, the team report on a treadmilling motion of the bacterial cytoskeleton which they have discovered is critical for division.
In the longer term, this study could open up novel antibiotic targets as it may be possible to develop new drugs that specifically inhibit this motion. This would be similar to how the chemotherapy drug, taxol, suppresses the motion of the cytoskeleton in cancer cells.
Bacterial cell division as target
The rise of antibiotic resistant bacteria threatens to return us to an era where even an infection from a minor scratch can kill.
One key antibiotic target is the process of bacterial cell division. However, for decades this process has been mysterious because cell division occurs on the nanoscale, invisible to conventional microscopy.
Teams from across the world, at Newcastle University in the UK, Harvard and Indiana Universities in the USA, and TU Delft in the Netherlands, worked together to use the latest imaging techniques to reveal the elusive mechanistic principles of bacterial cell division.
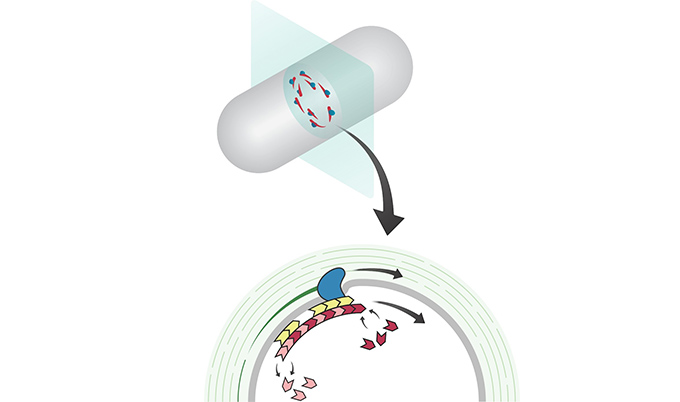
Bacteria divide against high internal pressure, which is like trying to cut an inflated balloon in two without bursting it. A ring of protein filaments forms around the future division site, and enzymes associated with this ring build a new crosswall that cleaves the bacteria in half. But what has remained completely mysterious is how these proteins work together as a single nanoscale machine to cut the bacterial balloon skin, cell wall, in two.
Researchers worked to track the dynamics of the newly inserted cell wall that divides the cell, the enzymes that build this material, and the underlying dynamics of the cytoskeletal protein filaments that orchestrate the whole process. They began by examining the motion of FtsZ, a cytoskeletal filament that is required for cytokinesis in bacteria and is related to the tubulin cytoskeletal protein found in eukaryotic cells.
Speed of filament motion critical
Using high-resolution microscopy techniques, they found that FtsZ filaments move around the division site, traveling around the division ring. They imaged the motion of individual cell wall synthesis enzymes, and saw that the synthesis enzymes ride on FtsZ filaments, building new cell wall as they travel around the division site. This causes the cell wall to be synthesized in discrete sites that travel around the division site during cytokinesis, a process which they were able to observe directly by using dyes that label the bacterial cell wall. Using a variety of experimental techniques, they were able to speed up or slow down how fast FtsZ rotated around the cell.
Dr Seamus Holden, from the Institute for Cell and Molecular Biosciences at Newcastle University and one of the senior authors of the study, said: “Strikingly, we found that the speed of FtsZ filament motion determines how fast the cell can divide. When FtsZ moves more rapidly, the cell wall is built more quickly, and cytokinesis happens faster. This shows that the motion of FtsZ is the critical overall controller of cell division.”
One challenge that the scientists faced was trying to look at the division proteins in actively dividing cells. At the earliest stages of division, it was possible to image division protein organization because the proteins in the partially assembled ring are sparsely distributed. However, a new strategy was required to measure how the dense protein network of actively dividing cells was organized.
Scientists normally immobilize bacteria flat on a microscope slide, and image at them from underneath, but unfortunately this places the division ring side-on, obscuring the motion and organization of division proteins. However, researchers had a trick up their sleeves - by using nanofabrication technology originally developed to manufacture computer chips, they were able to develop tiny gel nanocages to trap bacteria in an upright position.
Dr Holden added: “By trapping individual bacteria in sculpted nanocages, we were able to rotate the cell division ring so that it was fully visible on our high resolution microscope. This revealed the dynamic motion of FtsZ filaments as they travel around the entire division site.
“We are currently working on combining this approach with super-resolution microscopy to further increase resolution 10 to 20-fold and finally image the organization of individual cell division proteins and filaments.”
Together, these results revealed the basic mechanistic principles of bacterial cell division: that the building of the division crosswall is orchestrated by moving cytoskeletal filaments. Previously, the cytoskeleton was thought to serve as a static scaffold, recruiting other molecules and perhaps exerting some local force to divide the cell. This new work demonstrates that all the components of cell division are in constant, controlled motion around the division site, driven by the fundamental dynamics of the cytoskeleton.
In the longer term, this study could open up novel antibiotic targets. Based on the discovery that the treadmilling motion of the bacterial cytoskeleton is critical for division, it may be possible to develop new drugs that specifically inhibit this motion, similar to how the chemotherapy drug taxol suppresses the motion of the cytoskeleton in cancer cells.
Reference
Treadmilling by FtsZ Filaments Drives Peptidoglycan Synthesis and Bacterial Cell Division.
Alexandre W. Bisson Filho, Yen-Pang Hsu, Georgia R. Squyres, Erkin Kuru, Fabai Wu, Calum Jukes, Yingjie Sun, Cees Dekker, Seamus Holden, Michael S. VanNieuwenhze, Yves V. Brun, and Ethan C. Garner



